
BIOMOD Team Sendai
Overview
・Background
Macrophages are responsible for the “phagocytic process,” in which they autonomously
recognize and engulf pathogens or foreign substances. When they detect target bacteria or
foreign materials, macrophages enclose them in their cell membrane, internalize them, and
subsequently degrade and neutralize them inside the cell. This sequence of processes
constitutes a highly sophisticated biological system composing three steps: 1. specific
recognition, 2. signal transduction, and 3. active uptake.
DNA has attracted attention as a useful material for artificially reproducing such advanced autonomous functions.
DNA, a molecule used for encoding genetic information in cells, can be utilized
beyond information storage by leveraging its specific base-pairing and self-assembly
properties. These properties enable the construction of diverse nanostructures and
functional materials. One example of this is the DNA hydrogels. DNA hydrogels have
promising applications in fields such as sensing and drug delivery. By integrating the
diverse functions realized through DNA sequence design technologies could enable the
construction of highly autonomous systems analogous to macrophages.
・Problem
While DNA hydrogels offer excellent biocompatibility and programmability, their integration
with advanced technologies such as DNA computing has been challenging due to the
significant gap in operating concentrations between the hydrogel and the DNA computing.
・Goal
In this study, we set the goal of developing a functional DNA hydrogel, named “Hungry Gel,” that mimics the phagocytic behavior of macrophages and autonomously detects and engulfs target bacteria.
Idea
In order to reconstruct the phagocytic process of macrophages as a DNA-based system, we first broke down the macrophage mechanism into three abstract functional modules and reinterpreted each in a way that could be implemented using DNA molecules.
1. First, we achieved recognition by exposing specific sequences as a result of reaction with the target.
2. Second, we achieved signal transduction by converting the exposed signals through chemical amplification that produces many product molecules from trigger molecules.
3. Third, we achieved uptake of the target by locally degrading the material structure in response to the amplified signal, thereby recreating the engulfing behavior of macrophages through structural dissolution of the DNA hydrogel.
Through this functional decomposition, we defined the phagocytic process as a sequence of three independent modules: “recognition → signal transduction → uptake,” each implemented as interactions between DNA molecules.
Solution
We implement the three functional modules, which we abstracted above, as a concrete DNA-based system: DNA aptamers, entropy-driven circuits, and DNA hydrogels.
1. Module for recognizing targets and converting information
This module employs DNA aptamers that specifically bind to molecules on the surface of
target bacteria. Upon binding to the target, the aptamer undergoes a conformational
change, exposing a single-stranded DNA which triggers the amplifier.
2. Module for amplifying and converting signals
This module utilizes an entropy-driven circuit, which is driven by the trigger sequence
exposed through target recognition in the previous module, which act as the catalyst
sequence. The circuit consists of a solution containing three DNA components; substrate,
fuel, and catalyst. The substrate reacts with the catalyst to release output DNA while
consuming the fuel but leaving the catalyst. When the catalyst DNA exposed by the
aptamer is appeared, the reaction cycle initiates, producing a large amount of single-
stranded DNA called “output”.
3. Module for uptaking targets by self-dissolution of DNA hydrogel
This module employs a DNA hydrogel composed of four-branched DNA motifs as its
network components. This hydrogel binds adjacent motifs through two types of sticky ends
of different lengths.The short sticky ends are used for binding between motifs, while the long sticky ends are designed to have
sequences complementary to the output DNA. This design allows the output DNA to
preferentially bind to the long sticky ends, results in releasing the bond between the motifs
via toe-hold mediated strand displacement (TMSD), thereby dissolving the hydrogel's
network structure.
Based on these concepts, the system flow is illustrated in Fig. 1.
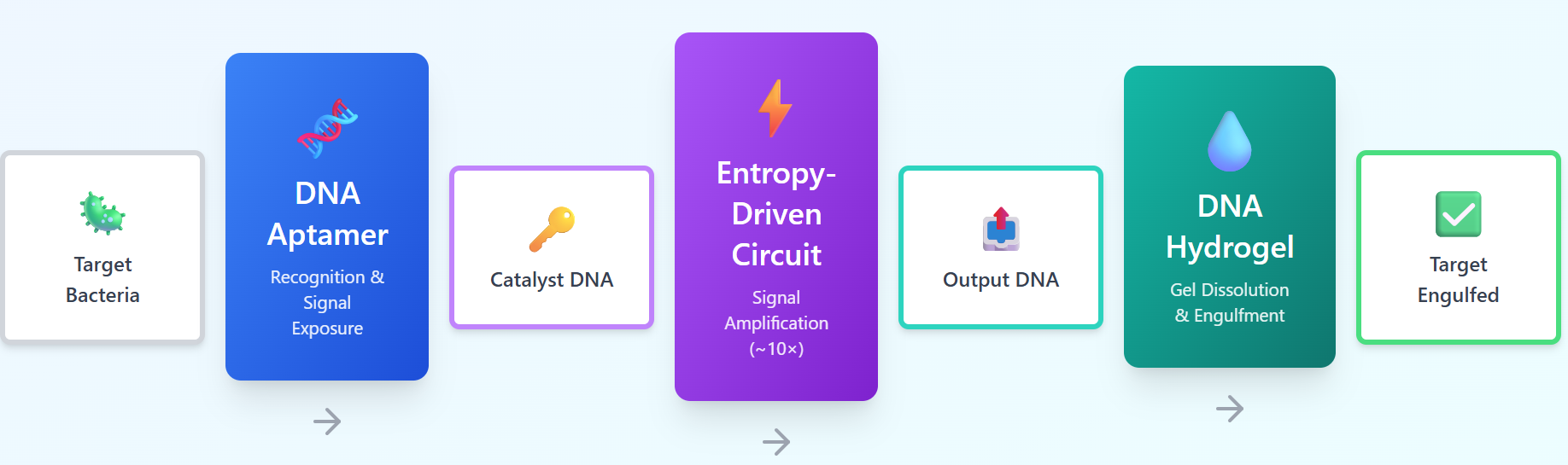
Fig.1 Overview of the Hungry Gel system
Milestones
We have established the five milestones as shown in Fig.2 The achievement level of this project is evaluated by these milestones.

Fig.2 Milestones of this project

